The UniBioPAN is a powerful tool designed to facilitate the analysis and prediction of bioactive peptide activity. The subsequent text offers a comprehensive elaboration of the user manual
The UnBioPAN Web Server User Manual
The UniBioPAN web server facilitates the prediction of bioactive peptides through a streamlined and user-friendly interface.
Users can efficiently navigate the prediction process by adhering to the following steps (Figure 1):
(1) Sequence Input (①): Enter FASTA-formatted sequences of the bioactive peptides you wish to predict. This interface allows for up to ten sequences to be submitted simultaneously.
(2) Threshold Selection (②): A threshold value can be adjusted to influence the prediction's success rate for positive identifications. A higher threshold strengthens the prediction for confirmed actives, while a lower threshold might identify more sequences as potentially active.
(3) Activity Selection (③): Choose the specific activities you want to predict. Selecting "all activity" will generate predictions for 31 distinct bioactive peptide categories.
(4) Task Submission (④): Submit your prediction task. Following submission, please be patient. The results will populate on this webpage upon completion. To prevent server overload, avoid resubmitting tasks. If the task fails, you may retry.
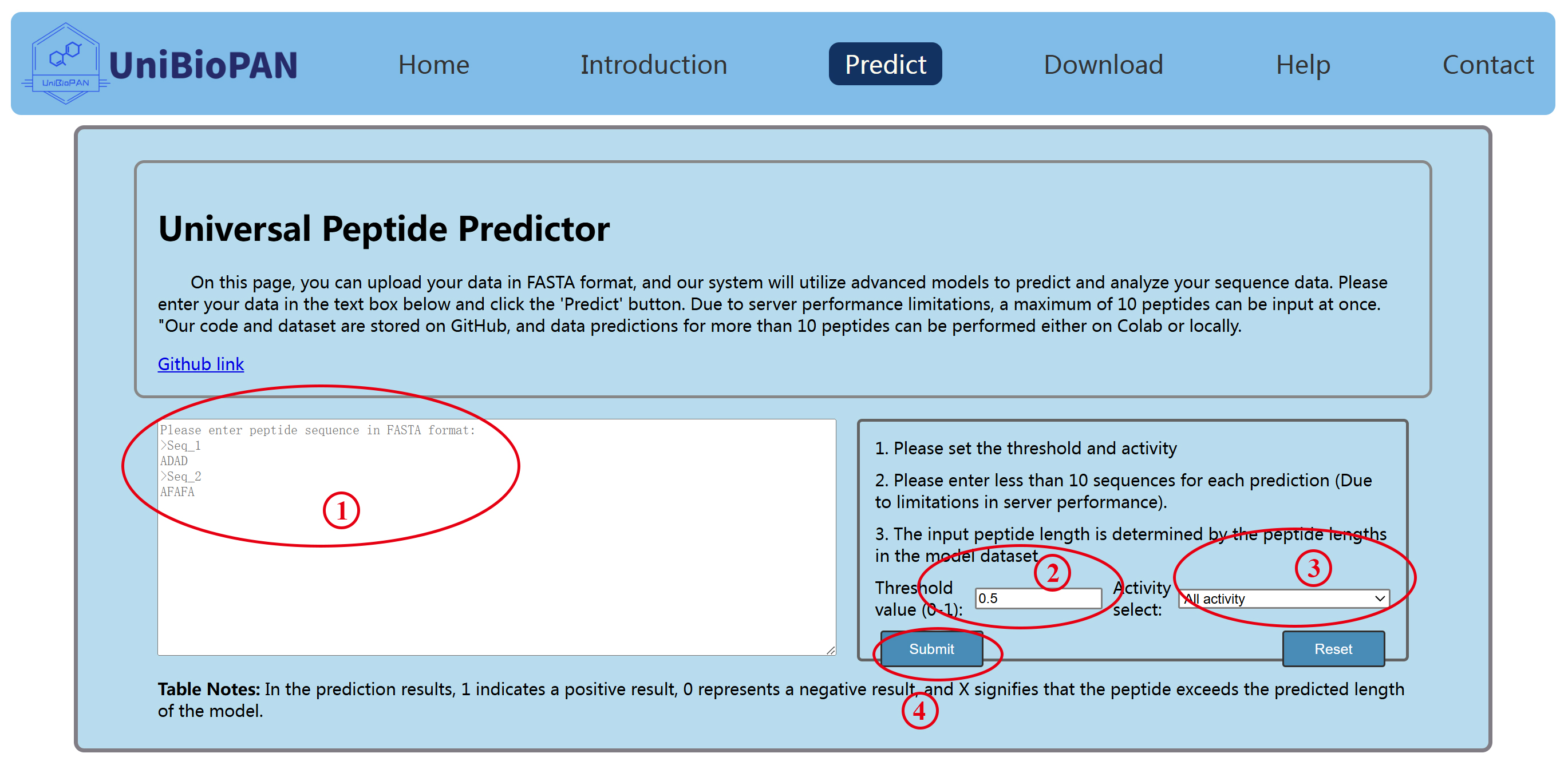
Figure 1. UniBioPAN bioactive peptide prediction interface
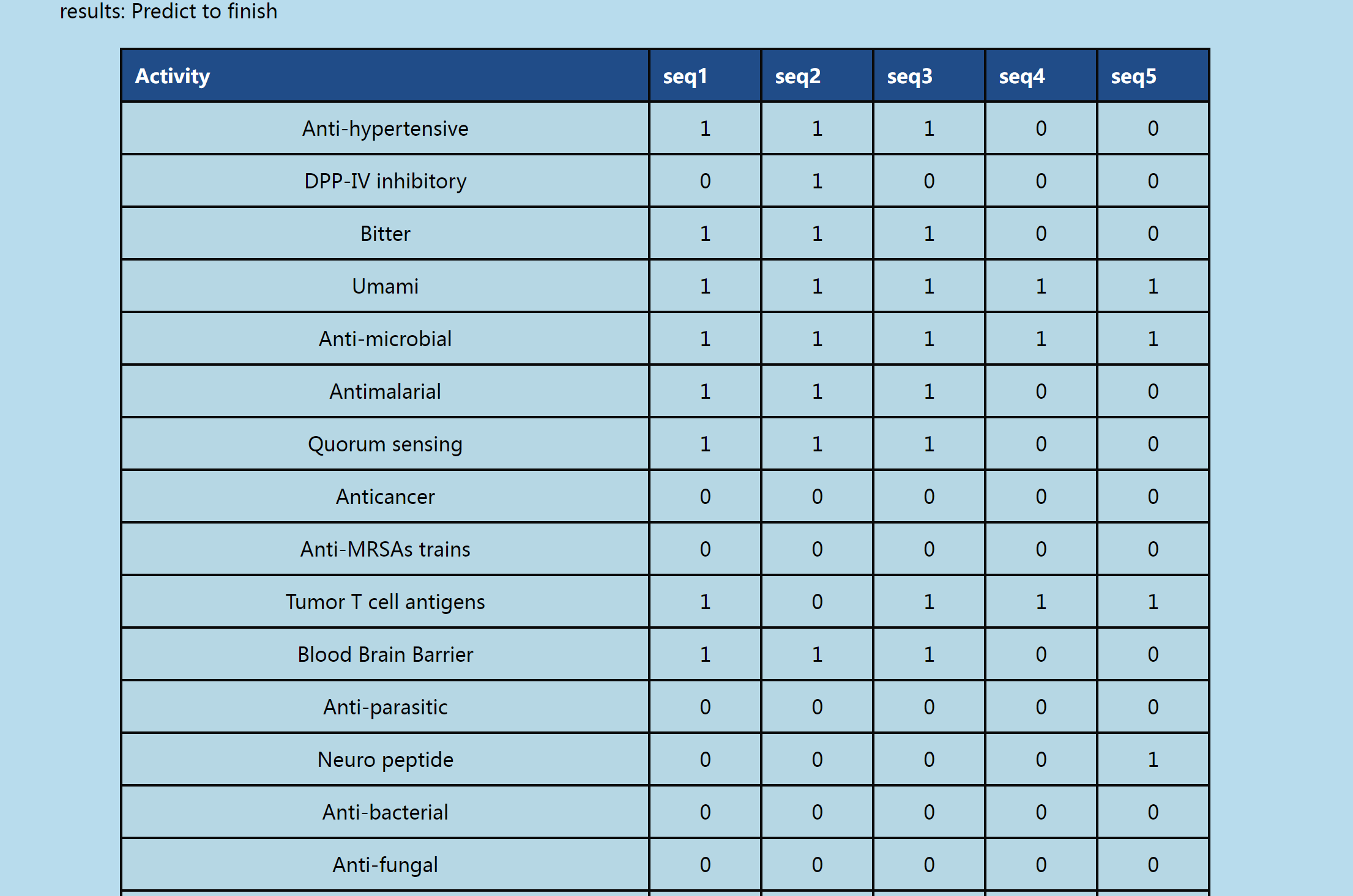
Figure 2. UniBioPAN bioactive peptide prediction results interface
This figure illustrates the interpretation of the prediction results generated by the UniBioPAN web server. The results are presented in a tabular format, with each row corresponding to an individual peptide sequence.
Key Interpretation Points:
(1) Positive Result: A value of 1 in the prediction result column indicates that the peptide is predicted to be bioactive.
(2) Negative Result: A value of 0 in the prediction result column indicates that the peptide is predicted to be inactive.
(3) the label is "x": An "x" in the prediction result column signifies that the peptide sequence either exceeds the maximum length limit for the model or contains non-standard amino acids.
It is crucial to note that the UniBioPAN web server provides predictions based on computational models and should not be considered definitive. Experimental validation is always recommended to confirm the bioactivity of peptides.
UniBioPAN's User Manual for Google Colab
Google Colab offers a free cloud-based platform
for training and predicting machine learning models. It is an ideal choice for researchers with limited hardware
esources to learn and apply deep learning techniques.
To embark on a new model training or prediction task, follow these comprehensive steps:
(1) Download files:
gather the three files depicted in Figure 3 into a single directory. Download link
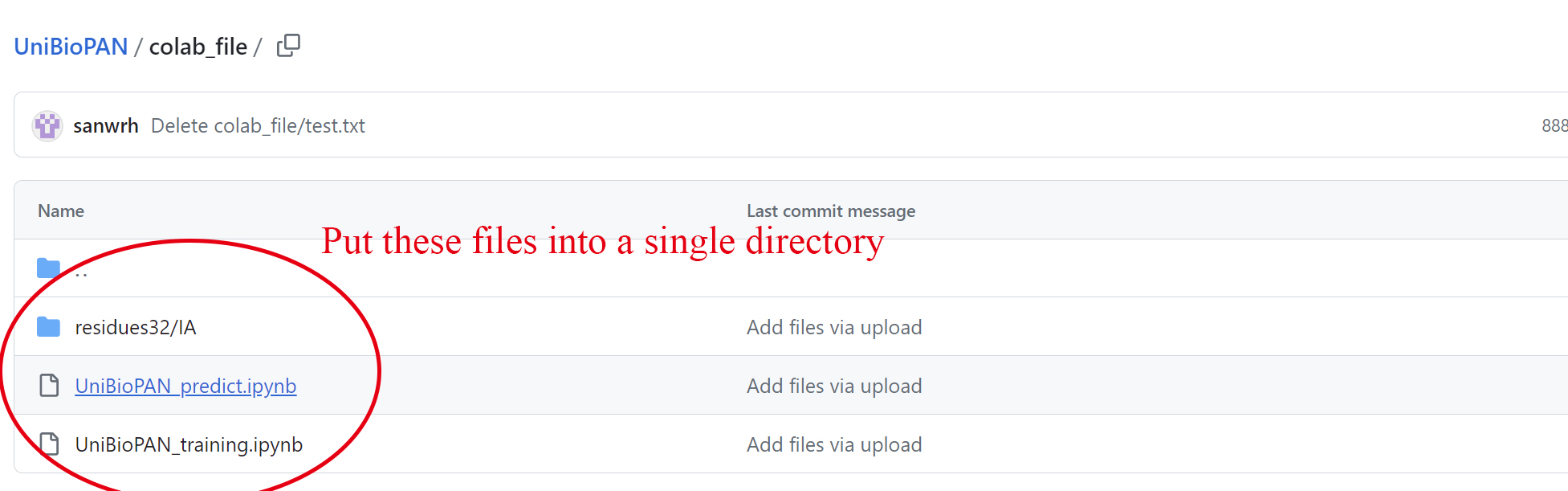
Figure 3. Retrieve colab files from github
(2) Environment setup and directory switching: execute the code displayed in the Figure 4 cell to install the required environment.
Subsequently, switch your working directory to the path where the three files from Step 1 are stored.
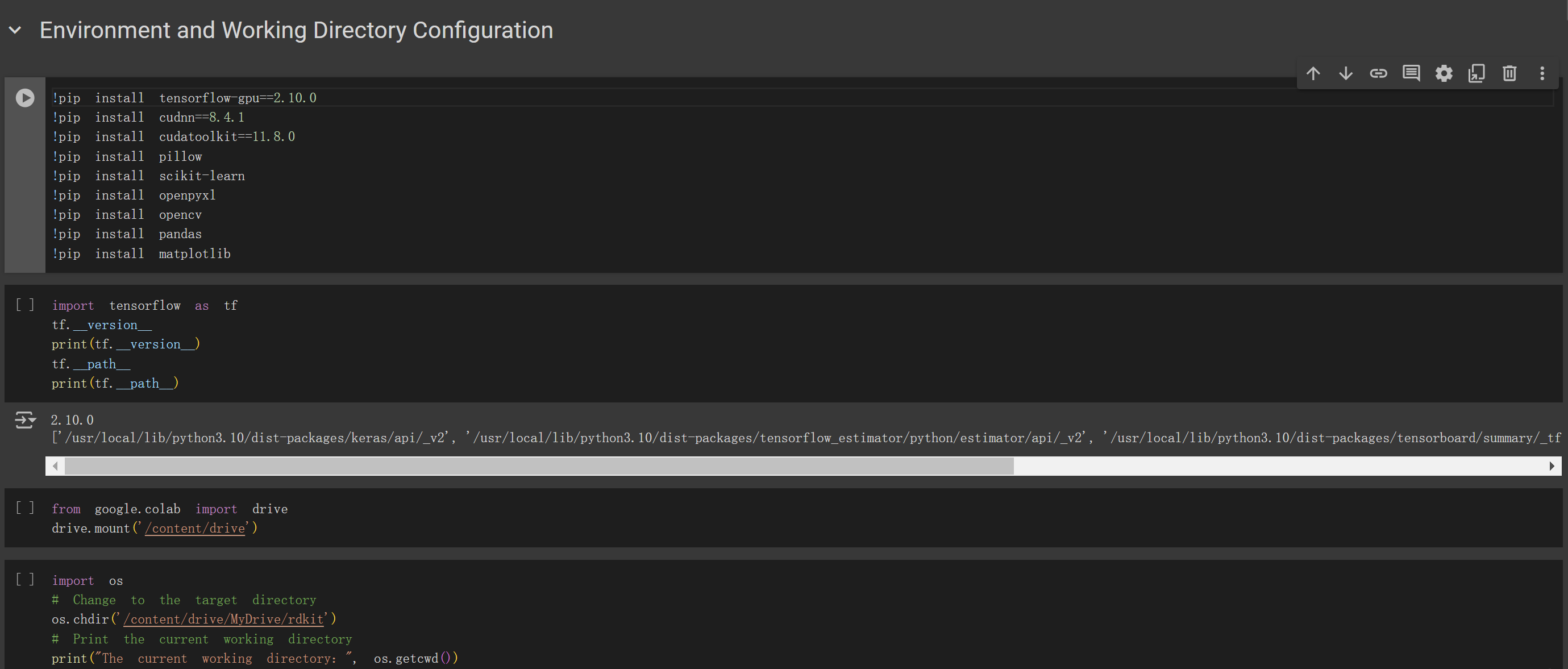
Figure 4. Environment setup and directory switching
(3) Parameter configuration and model training: configure the parameters according to the instructions provided in Figure 5.
Run the entire code to train the model. Upon completion, the trained model's performance metrics will be saved in the
designated XLSX file.
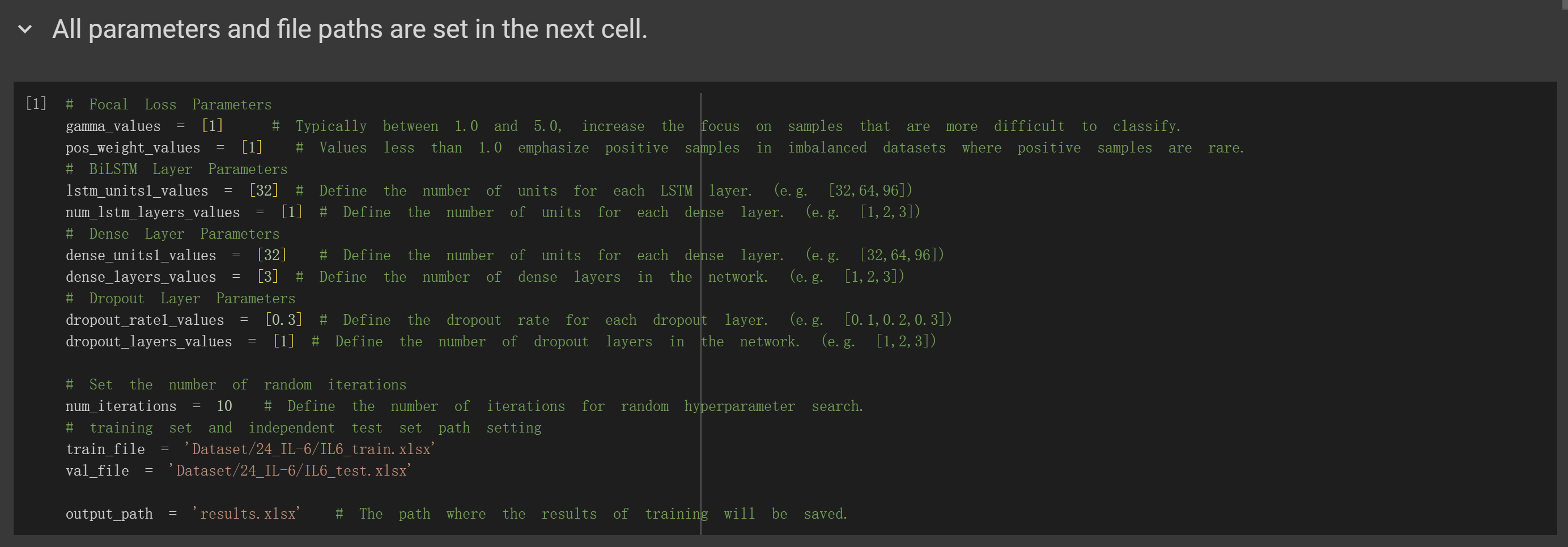
Figure 5. Parameter configuration and model training
(4) Prediction: based on the results obtained in Step 3, identify the model path with the optimal performance. Input the path to this model and the path to the prediction file in the code shown in Figure 6. The prediction file can be in FASTA, CSV, or XLSX format. Refer to the example files provided in the 'example' directory.
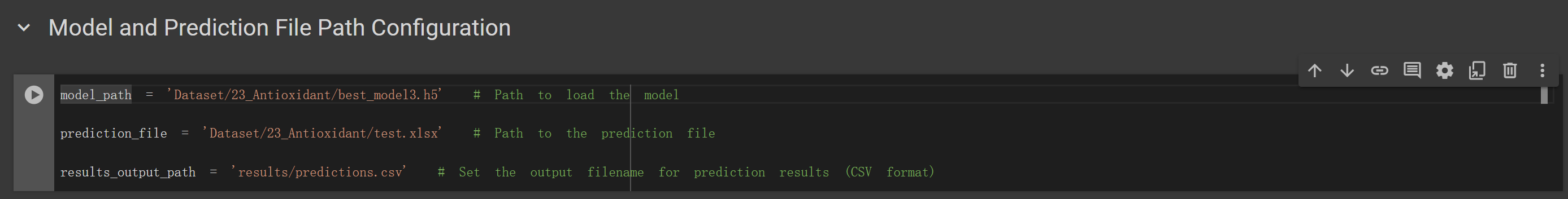
Figure 6. Prediction Parameter Settings Interface
UniBioPAN's User Manual for python script
UniBioPAN script offers a more flexible way to use it on your local computer.
To use UniBioPAN script, you can follow these steps:
(1) First, create a conda environment. You can download the enviroment.yml file from GitHub and use the following command to set up the environment:
> conda env create -f environment.yml --prefix /path/to/tensor
(2) After the environment is set up, you need to download the three files shown in Figure 7 to the same folder.
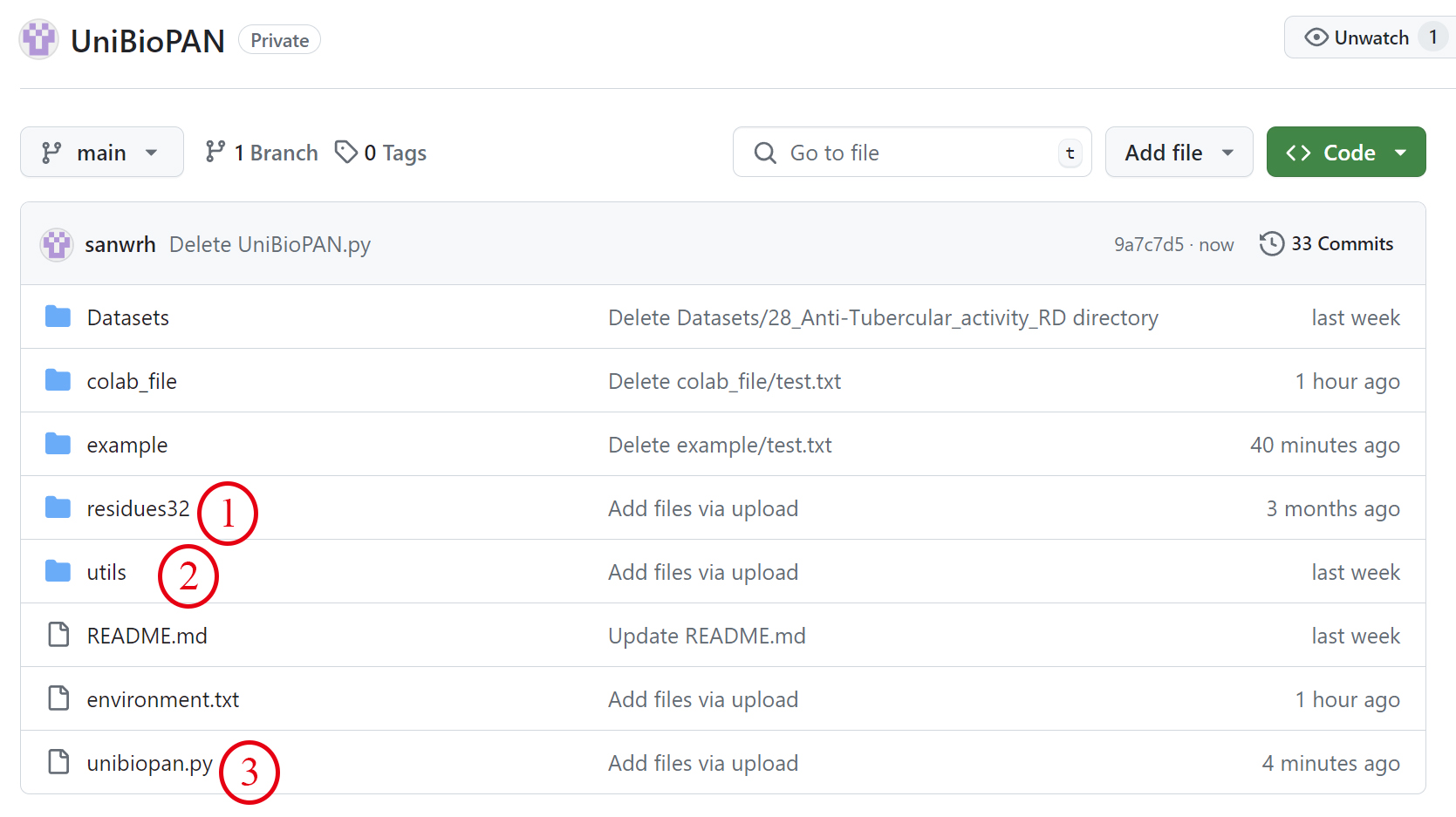
Figure 7. Download UniBioPAN script
(3) Use the command:
> python unibiopan.py -h
to view the script's help information (Fiugre8).
(4) UniBioPAN script has two modes: training mode and prediction mode.
To train a model, use the command:
> python unibiopan.py -t -tf Dataset/train.xlsx -ef Dataset/test.xlsx
The -tf (training file) and -ef (independent test file) arguments are required, and the file format must be xlsx.
To make predictions, use the command:
> python unibiopan.py -p -lp model/best_model.h5 -pf predict/input.xlsx
The -lp (load path for the model) and -pf (prediction file) arguments are required. The input file format can be fasta, csv, or xlsx.
Figure 8. UniBioPAN script help interface